Track properties
type
Requried. type specifies the track type, currently supported track
types:
- bigWig
- bedGraph
- dynseq
- methylC
- categorical
- hic
- bed
- bigbed
- refbed
- repeatmasker
- geneAnnotation
- genomealign
- longrange
- bigInteract
- qBED
- matplot
- snp
- g3d
- ruler
type is case insensitive.
name
Requried. name specifies the track name used internally by the browser. It is also displayed as the track legend if no label speficied. Value can be any string.
label
Optional. label specifies the track legend displayed in the browser. It overrides the name arrtibute. Value can be any string.
url
Requried. url contains the URL to the track file and needs to be HTTP or HTTPS location string.
A url is requried for all the tracks in binary format. Gene annotaion tracks, like refGene, do not need a url as they are stored in the Mongo database. Additional annotation tracks, such as the ruler track, also do not need a url.
Each user-provided url must link to a publically available website, without password protection, so that the browser can read in the file.
url can use a relative child path to the datahub url, say you have a file a.bigWig with your datahub http://your.host/your.hub.json, when you add the track entry for a.bigWig, the url can be either http://your.host/a.bigWig or just a.bigWig.
showOnHubLoad
Optional. If specified to true, the track will be displayed when hub is loaded. Default value: false.
metadata
Optional. An object specifying the metadata of the track.
In this basic example the value of each metadata term is a string. :
"metadata": {
"sample": "bone",
"assay": "MRE"
}
This example public Roadmap data hub has more complex metadata definitions and makes use of a list of strings to build a hierarchical structure. :
{
"url": "https://egg.wustl.edu/d/hg19/GSM997242_1.bigWig",
"metadata": {
"Sample": [
"Adult Cells/Tissues",
"Blood",
"Other blood cells",
"CD4+_CD25-_Th_Primary_Cells"
],
"Donor": [
"Donor Identifier",
"Donor_332"
],
"Assay": [
"Epigenetic Mark",
"Histone Mark",
"H3",
"H3K9",
"H3K9me3"
],
"Institution": [
"Broad Institute"
]
},
"type": "bigwig",
"options": {
"color": "rgb(159,0,72)"
},
"name": "H3K9me3 of CD4+_CD25-_Th_Primary_Cells"
}
The list of metadata is ordered from more generic to more specific and helps build the facet table hierarchy making the search and filter functions in track table easier.
details
Optional. If you want to add more information for each track then the details attribute is helpful. After right clicking on the track you can click More Information and see the details, url, and metadata for each track in the dropdown menu. :
"details": {
"data source": "Roadmap Project",
"date collected": "May 7 2016"
}
queryEndPoint
Optional. Most time this parameter will be used with geneAnnotation track. When users deal with custom genome, or genome not listed in NCBI or Ensembl database, gene search link would not work, so in such case, user can specify a custom database to query detailed information. For example, for some trypanosome genome, gene search should be queried through TriTryDB, we can define the track like this then:
{
type: "geneAnnotation",
name: "gene",
label: "TriTrypDB genes",
genome: "TbruceiLister427",
queryEndpoint: { name: "TriTrypDB", endpoint: "https://tritrypdb.org/tritrypdb/app/search?q=" },
}
options
Optional. All track render options are placed in an object called options. This object can have the following properties:
color
color is used to define the color for each track. A color name, RGB values, or hex color code can be used. For more about color name or RGB please see https://www.w3schools.com/css/css_colors.asp.
color2
color2 is used to define the color for negative values from the track data. The default is the same as color.
backgroundColor
backgroundColor defines the background color of the track.
height
height controls the height of the track which is specified as a number and displayed in pixels.
ensemblStyle
currently for [bigwig] track, specify ensemblStyle option to [true] can enable data with chromosome names as 1, 2, 3...work in the browser
yScale
yScale allows you to configure the track's y-scale. Options include auto or fixed. auto sets the y-scale from 0 to the max value of values in the view region for a given track. fixed means you can specify the minimal and maximal value.
yMax
yMax is used to define the maximum value of a track's y-axis. Value is number.
yMin
yMin is used to define the minimum value of a track's y-axis. Value is number.
If you need the track to be in fixed scale, you need to specify yScale to fixed besides of set yMax and yMin.
group
Numerical tracks can be grouped to same group, tracks from same group will share y-axis scale settings. For example, when 2 tracks are in one group, the y-axis will both set to max value of current views of both tracks. Users can find one example data hub with group settings from here: https://wangftp.wustl.edu/~dli/test/a-group.json
scoreScale/scoreMax/scoreMin
These options work similar as yScale/yMax/yMin, but these are for interaction tracks.
colorAboveMax
colorAboveMax defines the color displayed when a fixed yScale is used and a value exceeds the yMax defined.
color2BelowMin
color2BelowMin defines the color displayed when a fixed yScale is used and a value is below the yMin defined.
displayMode
displayMode specifies display mode for each tracks. Different tracks have different display modes as listed below.
| type | display mode |
|---|---|
| bigWig | auto, bar, heatmap |
| bedGraph | auto, bar, heatmap |
| geneAnnotation | full, density |
| HiC | arc, heatmap, flatarc, square |
| genomealign | rough, fine |
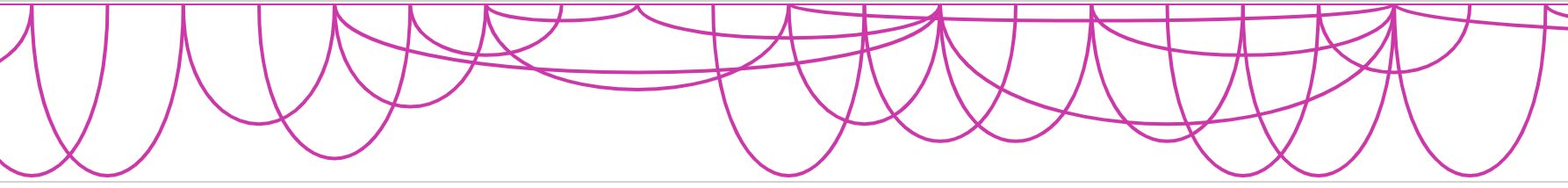
flatarc mode
For interaction track. flatarc mode is like arc mode, sometimes the curve would be displayed flatter, in fact it's a cubic curve.
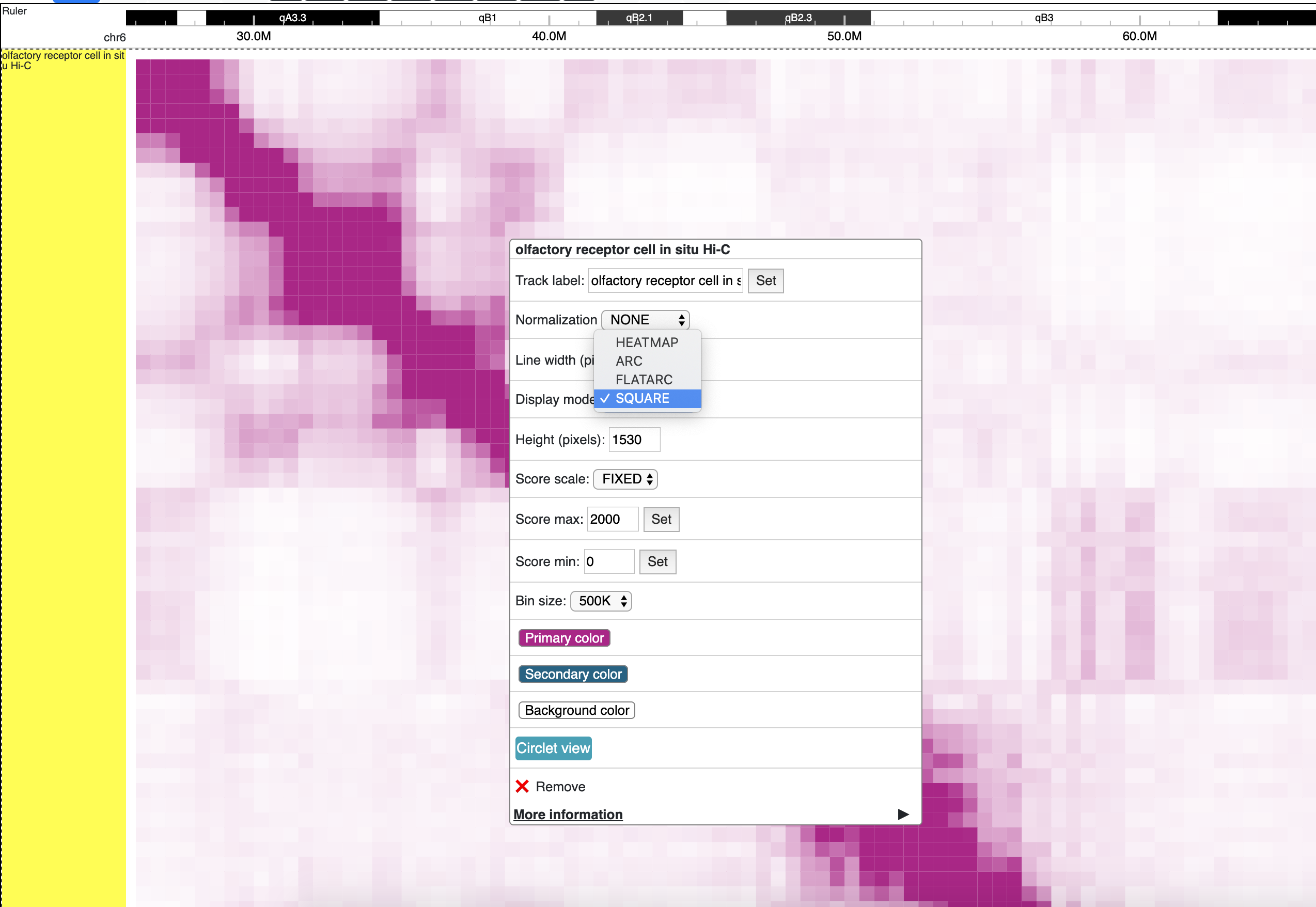
square mode
For interaction track. square mode gives JuiceBox style like view for HiC maps.
aggregateMethod
At high zoom-out level when 1 on-screen pixel spans >1bp, the underlying track data needs to be summarized into a single value for browser display. aggregateMethod is used to control how the data is summarized. Supported values include: MEAN, SUM, COUNT, MAX, MIN. Default value is MEAN.
smooth
smooth option allows you to smooth the graph of a quantitative track using window mean values. The browser will use the mean values from region [current_position - smooth, current_position + smooth]. Default value is 0 (no smooth applied).
maxRows
maxRows options controls the number of rows for the annotation track, like a geneAnnotation track.
hiddenPixels
For annotation tracks, when an element spans less than [hiddenPixels] in the screen, this item will not be displayed. Default value is 0.5 pixel. Set to 0 will display all elements.
isCombineStrands
For methylC tracks, isCombineStrands will specificy if the strands should be combined true or not combined false. We recommend combining stands for viewing CpG methylation, but leaving strand information for non-CpG methylation.
depthFilter
For methylC tracks a depthFilter can be set to filter out any bases with less than the depth(coverage) specified.
depthColor
For methylC tracks specify a depthColor for the depth line that overlays the bars.
maxMethyl
For methylC tracks specify the y-axis max (for both strands) using maxMethyl. Options range from (0-1].
zoomLevel
For bigWig track only. bigWig files usually contain multiple resolutions, when viewing a large region, the Browser usually fetches a lower resolution for faster response, user can change this behaviour by changing this option.
The example below first show viewing a bigWig track in a big region with AUTO zoom level, you can see the data is pretty flat, when we change zoom level to 0, 1, etc, we can see more details from the data, but takes more time to load.
Automatical zoom level:

Right click, change zoom level to 0: (can also setup in data hub under options)
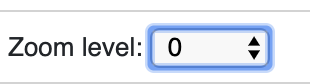
View changed after change zoom level to 0:

alwaysDrawLabel
For bed and categorical tracks only. Usually for each bed and [`categorical] item in those tracks, the label are only drawn only when there are enough space inside the item block, by specificy this option to [true], the label will always be drawn in the screen.